Experimenting with Cellular Automata
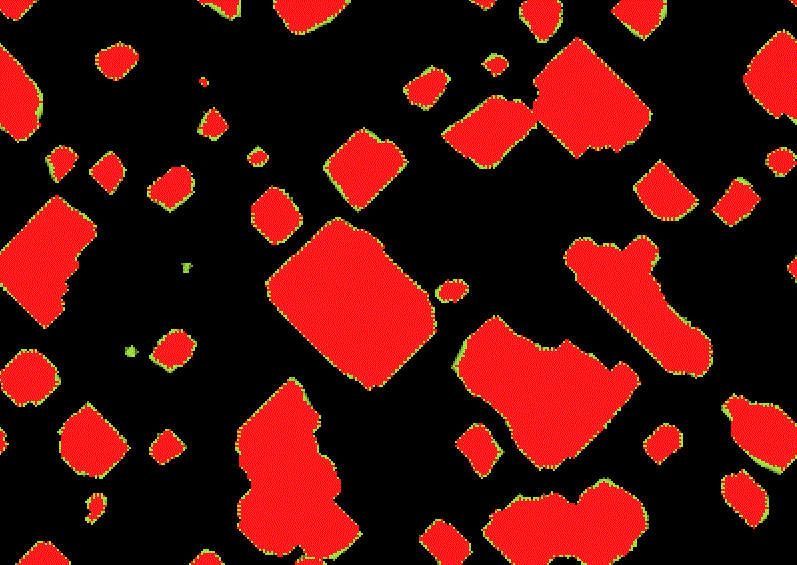
Diamoeba rule using S5678/B35678
Some time ago I experimented with cellular automata (CA), the most famous of which is Conway’s Game of Life. Cellular automata are a set of rules based systems for generating life-like simulations.
There are a number of applications to cellular automata, including:
- Procedural generation in games such as cave generation.
- Modelling the spread of forest fires.
- Random number generation.
The Game of Life automaton can even be used to simulate itself.
I created my own app to experiment with some of the different CA rules. I created this in C with SDL, a low level game development library. Later, I ported this to Emscripten which has built-in support for SDL, to compile this to run in a browser. A demo of this is available below.
The core algorithm is fairly simple. For each cell in the grid we loop over the immediate neighbourhood and count the cells alive:
for (x = 0; x < width; x++)
{
for (y = 0; y < height; y++)
{
int neighbours = 0;
int idx = 0;
uint8_t current;
uint8_t next;
for (xn = -1; xn <= 1; xn++)
{
for (yn = -1; yn <= 1; yn++)
{
if (xn == 0 && yn == 0) continue;
// Neighbourhood index, wrap to the opposite side at edges.
idx = mod((x + xn), width) + width * mod((y + yn), height);
neighbours += in[idx];
}
}
...
Then the next state is determined by looking at the rules look-up-table. The pixel data is then written, I have added a ‘dying’ state for cells that have just changed from alive to dead to customise the look:
...
current = in[idx];
next = rules[current][neighbours];
out[idx] = next;
if (next)
{
pixels[idx] = alive;
}
else if (current != next)
{
pixels[idx] = dying;
}
else
{
// Shift out alpha bits to fade out dying cells
pixels[idx] = ((pixels[idx] & AMASK) << 1) | (~AMASK & pixels[idx]);
}
}
}
This implementation is the naive or brute-force method though. This could be optimised by avoiding the repeated calculations of the neighbourhood indices and using a pre-calculated look up table, you would then be able to use a single for-loop rather than the four nested loops.
Additionally, the naive algorithm wastes time looking at areas that are not changing. This could be improved by keeping track of all the cell transitions and looking only at the cells adjacent to the transitions. Even so, the naive approach has no problem running at 60FPS in a modern browser.
The full source is available at github.
Demo
Use the controls on the right to changes the CA rules applied and to adjust the colours, (changing the rules currently requires a restart). You can find a list of interesting rules here.